We are ready to help you overcome the challenges of producing and developing innovative biotechnological products and advancing scientific research

RMSD analysis indicates the stability of the protein structure throughout the Molecular Dynamics simulation. Minor changes in the RMSD value suggest the overall structure is stable, while large fluctuations may indicate the occurrence of major structural changes. This parameter is used to assess the agreement of the structure with the initial model.

SASA is the surface area of a protein molecule that is in contact with the solvent (such as water). This parameter is used to investigate structural changes, ligand interactions, and assess the stability of hydrophilic and hydrophobic regions. Significant changes in SASA can indicate changes in the folding or unfolding of the structure

Ligand RMSD analysis indicates the extent to which the ligand remained in its binding site or underwent displacement and conformational change during the simulation. A low RMSD value for the ligand suggests the ligand’s stability within its binding pocket on the protein.

RMSF is used to measure the fluctuation of each residue during the simulation. Regions with high RMSF often include loops or flexible regions, while areas with low RMSF indicate stable, structural parts of the protein, such as beta sheets or alpha helices.

MM/PBSA (Molecular Mechanics/Poisson-Boltzmann Surface Area) analysis is a common method used to calculate the binding free energy between a ligand and a protein. This analysis is performed as a post-processing step after the Molecular Dynamics simulation is complete and includes the sum of van der Waals, electrostatic, polar, and non-polar solvation energies. A high negative value (in absolute magnitude) for the binding free energy indicates a strong interaction between the ligand and the protein. This analysis is highly useful for assessing complex stability and comparing the binding efficiency of different ligands.
Energy Decomposition Analysis is a complementary step to MM/PBSA analysis that examines the contribution of each individual protein residue to ligand binding. This method identifies which amino acids play the most significant role in the stability of the protein-ligand complex
By identifying key residues that provide the greatest binding energy, this information can be utilized in drug design or protein engineering modifications. This analysis separately quantifies the electrostatic energies, van der Waals energies, and the solvation energy contribution for each residue
We are ready to partner with you in overcoming production challenges, developing innovative biotechnological products, and advancing scientific research


Energy Decomposition Analysis is a complementary step to MM/PBSA analysis that examines the contribution of each individual protein residue to ligand binding. This method identifies which amino acids play the most significant role in the stability of the protein-ligand complex
By identifying key residues that provide the greatest binding energy, this information can be utilized in drug design or protein engineering modifications. This analysis separately quantifies the electrostatic energies, van der Waals energies, and the solvation energy contribution for each residue

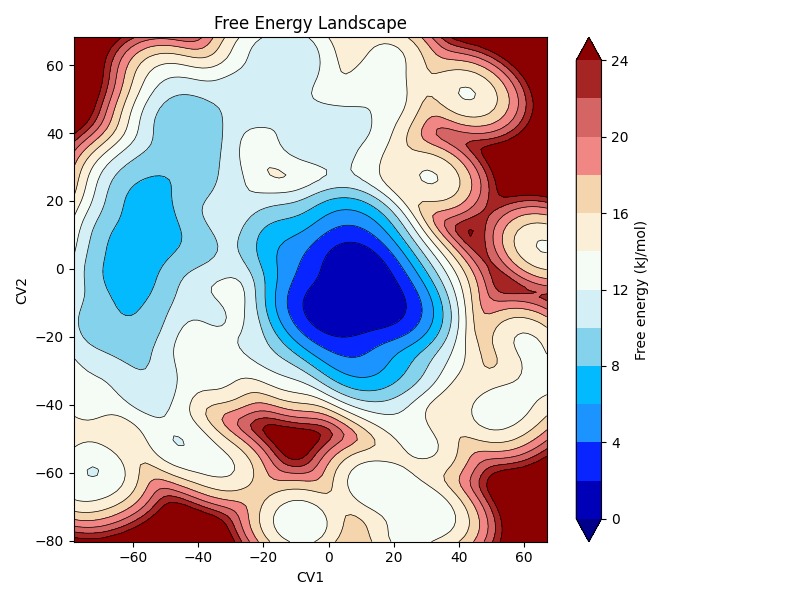
Our principal component analysis (PCA) service deciphers the dominant motions and conformational flexibility of proteins from simulation data. This approach reveals collective motions essential for enzymatic function, folding, and ligand binding, supporting deeper structural and dynamic insights for engineered proteins.

Dynamic Cross-Correlation Mapping (DCCM) analysis is used to examine the motional relationships between different regions of the protein. Regions with positive correlation move synchronously with each other, while negative correlation indicates movement in opposite directions. This analysis is useful for identifying functionally linked dynamic regions within the protein structure.
+98 912 836 0916
+98 930 144 1004
nimanezhad86@gmail.com


Custom-optimized rational enzyme engineering for your process requirements
©۲۰۲5 .neoenzyme all rights reserved